[의학통계방법론] Ch13. Data Transformations
Data Transformations
R 프로그램 결과
R 접기/펼치기 버튼
패키지 설치된 패키지 접기/펼치기 버튼
getwd()
## [1] "C:/Biostat"
library("readxl")
library("dplyr")
library("kableExtra")
엑셀파일불러오기
#모든 시트를 하나의 리스트로 불러오는 함수
read_excel_allsheets <- function(file, tibble = FALSE) {
sheets <- readxl::excel_sheets(file)
x <- lapply(sheets, function(X) readxl::read_excel(file, sheet = X))
if(!tibble) x <- lapply(x, as.data.frame)
names(x) <- sheets
x
}
13장
13장 연습문제 불러오기
#data_chap13에 연습문제 13장 모든 문제 저장
data_chap13 <- read_excel_allsheets("data_chap13.xls")
#연습문제 각각 데이터 생성
for (x in 1:length(data_chap13)){
assign(paste0('ex13_',c(1:4))[x],data_chap13[x])
}
#연습문제 데이터 형식을 리스트에서 데이터프레임으로 변환
for (x in 1:length(data_chap13)){
assign(paste0('ex13_',c(1:4))[x],data.frame(data_chap13[x]))
}
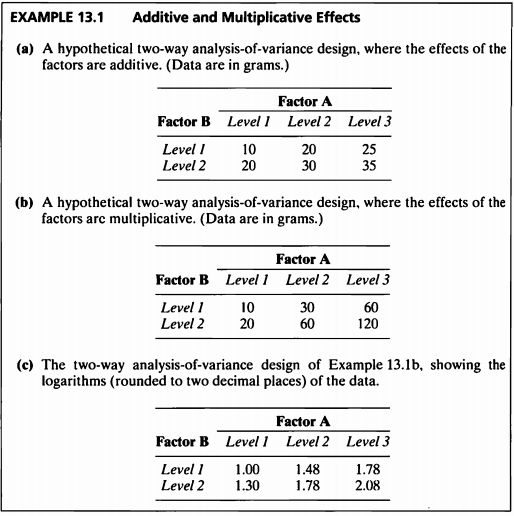
EXAMPLE 13.1
#데이터셋
ex13_1%>%
kbl(caption = "Dataset",escape=F) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam13_1.part | exam13_1.B | exam13_1.A | exam13_1.mean |
|---|---|---|---|
| 1. | 1 | 1 | 10.00 |
| 1. | 2 | 1 | 20.00 |
| 1. | 1 | 2 | 20.00 |
| 1. | 2 | 2 | 30.00 |
| 1. | 1 | 3 | 25.00 |
| 1. | 2 | 3 | 35.00 |
| 1. | 1 | 1 | 10.00 |
| 1. | 2 | 1 | 20.00 |
| 1. | 1 | 2 | 30.00 |
| 1. | 2 | 2 | 60.00 |
| 1. | 1 | 3 | 60.00 |
| 1. | 2 | 3 | 120.00 |
| 1. | 1 | 1 | 1.00 |
| 1. | 2 | 1 | 1.30 |
| 1. | 1 | 2 | 1.48 |
| 1. | 2 | 2 | 1.78 |
| 1. | 1 | 3 | 1.78 |
| 1. | 2 | 3 | 2.08 |
- 해당 데이터는 a)와 multiplicate된 데이터 b)와 b)의 로그변환된 데이터 c)로 구성되어 있으며
세개의 데이터로 Interaction plot을 그려보고자 한다.
Interaction plot
par(mfrow=c(1,3))
interaction.plot(ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.A,ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.B,ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.mean,bty="l",xlab="Factor A", ylab="mean", trace.label = "Factor B",main="Interaction plot of a)", las=1, type="b", pch=16, col=c("orange","#8f7450"))
interaction.plot(ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.A,ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.B,ex13_1[ex13_1$exam13_1.part=='b)',]$exam13_1.mean,bty="l",xlab="Factor A", ylab="mean", trace.label = "Factor B",main="Interaction plot of b)", las=1, type="b", pch=16, col=c("orange","#8f7450"))
interaction.plot(ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.A,ex13_1[ex13_1$exam13_1.part=='a)',]$exam13_1.B,ex13_1[ex13_1$exam13_1.part=='c)',]$exam13_1.mean,bty="l",xlab="Factor A", ylab="mean", trace.label = "Factor B",main="Interaction plot of b)", las=1, type="b", pch=16, col=c("orange","#8f7450"))
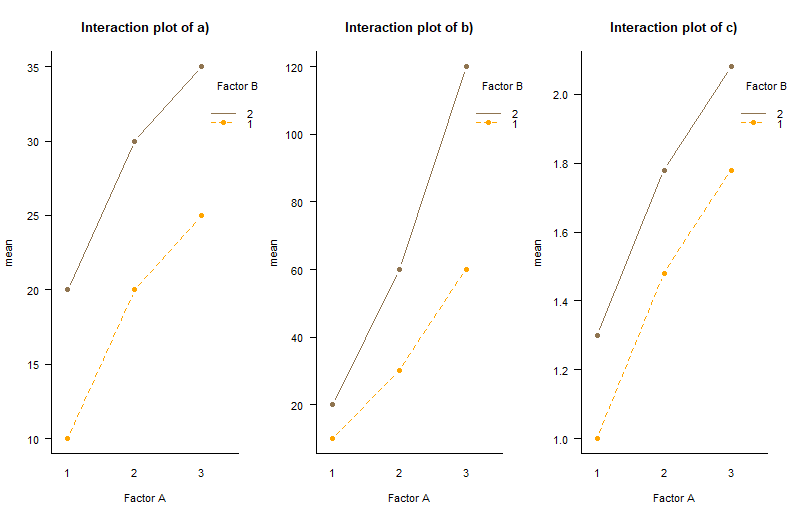
- 교호작용 그래프를 그려본 결과 위와 같은 결과가 나왔다.
- a), b), c)의 경우 Factor A가 level 3이고 Factor B가 level 2 일 때 최대의 평균을 가지며
그 중 b)를 로그변환한 자료인 c)의 교호작용 그래프를 보면 보다 조금 더 추세적으로 안정화된 교호작용 그래프가 그려짐을 볼 수 있다.
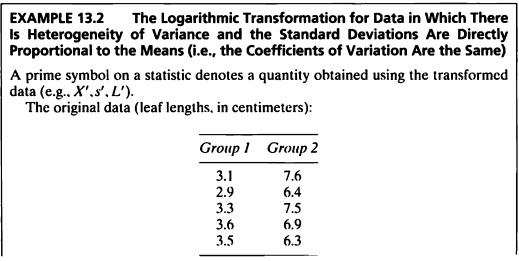
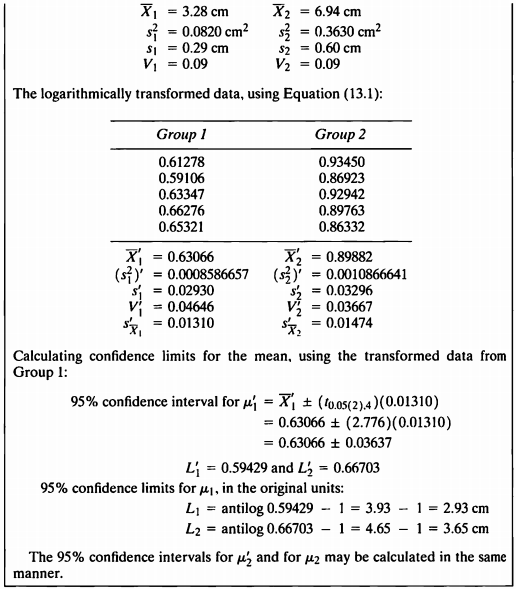
EXAMPLE 13.2
#데이터셋
ex13_2%>%
kbl(caption = "Dataset",escape=F) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam13_2.Group | exam13_2.Y |
|---|---|
| 1 | 3.1 |
| 1 | 2.9 |
| 1 | 3.3 |
| 1 | 3.6 |
| 1 | 3.5 |
| 2 | 7.6 |
| 2 | 6.4 |
| 2 | 7.5 |
| 2 | 6.9 |
| 2 | 6.3 |
- 해당 데이터를 사용하여 로그변환을 고려해볼 것이다.
- 로그변환은 분산이 동질적이지 않으면서 표준편차는 평균에 비례적인 경우 사용할 수 있다.
- original data를 로그변환한 데이터로 신뢰구간을 구하고, 그 값의 역로그변환을 하여 original data의 신뢰구간을 구하는 것도 볼 것이다.
Group1 <- subset(ex13_2$exam13_2.Y,ex13_2$exam13_2.Group==1)
Group2 <- subset(ex13_2$exam13_2.Y,ex13_2$exam13_2.Group==2)
ex13_2_split <- data.frame(Group1,Group2)
ex13_2_split%>%
kbl(caption = "Dataset of Example 13.2") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Group1 | Group2 |
|---|---|
| 3.1 | 7.6 |
| 2.9 | 6.4 |
| 3.3 | 7.5 |
| 3.6 | 6.9 |
| 3.5 | 6.3 |
\[\begin{aligned} X' = log_{10}(X+1) \end{aligned}\]로그변환
- 위 식을 사용하여 로그변환을 시도하겠다.
log_ex13_2 <- log10(ex13_2_split+1)
log_ex13_2%>%
kbl(caption = "Logarithmic transformation of Example 13.2") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Group1 | Group2 |
|---|---|
| 0.6127839 | 0.9344985 |
| 0.5910646 | 0.8692317 |
| 0.6334685 | 0.9294189 |
| 0.6627578 | 0.8976271 |
| 0.6532125 | 0.8633229 |
parameter <- c("Mean","Variance","Std","CV")
f13_2 <- function(x){
mean <- mean(x)
s2 <- var(x)
s <- round(sd(x),2)
V <- round(sd(x)/mean(x),2)
res <- list(mean,s2,s,V)
return(res)
}
Group_1_original <- f13_2(ex13_2_split$Group1)
Group_2_original <- f13_2(ex13_2_split$Group2)
origin <- data.frame(cbind(parameter,Group_1_original,Group_2_original))
origin%>%
kbl(caption = "Result of Original dataset") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| parameter | Group_1\_original | Group_2\_original |
|---|---|---|
| Mean | 3.28 | 6.94 |
| Variance | 0.082 | 0.363 |
| Std | 0.29 | 0.6 |
| CV | 0.09 | 0.09 |
parameter <- c("Mean","Variance","Std","CV","s_p","Lower CI","Upper CI","Original Lower CI","Original Upper CI")
log_f13_2 <- function(x){
mean <- mean(x)
s2 <- var(x)
s <- round(sd(x),4)
V <- round(sd(x)/mean(x),2)
s_p <- s/sqrt(length(x))
L1_log <- round(mean(x)-((qt(1-(0.05/2), length(x)-1))*s_p), 5)
L2_log <- round(mean(x)+((qt(1-(0.05/2), length(x)-1))*s_p), 5)
L1_origin=round(10^(L1_log)-1, 2)
L2_origin=round(10^(L2_log)-1, 2)
res <- list(mean,s2,s,V,s_p,L1_log,L2_log,L1_origin,L2_origin)
return(res)
}
Group_1_logtrans <- log_f13_2(log_ex13_2$Group1)
Group_2_logtrans <- log_f13_2(log_ex13_2$Group2)
logtrans <- data.frame(cbind(parameter,Group_1_logtrans,Group_2_logtrans))
logtrans%>%
kbl(caption = "Result of logarithmic transformation dataset") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| parameter | Group_1\_logtrans | Group_2\_logtrans |
|---|---|---|
| Mean | 0.6306575 | 0.8988198 |
| Variance | 0.0008585314 | 0.001086546 |
| Std | 0.0293 | 0.033 |
| CV | 0.05 | 0.04 |
| s_p | 0.01310336 | 0.01475805 |
| Lower CI | 0.59428 | 0.85784 |
| Upper CI | 0.66704 | 0.93979 |
| Original Lower CI | 2.93 | 6.21 |
| Original Upper CI | 3.65 | 7.71 |
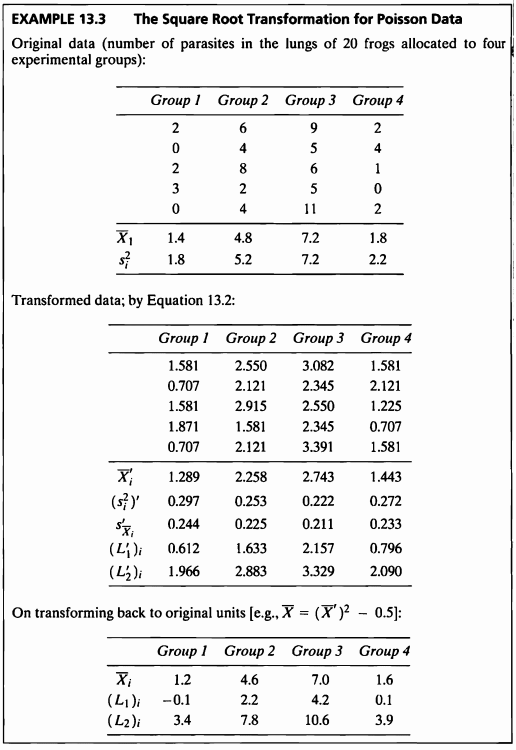
EXAMPLE 13.3
#데이터셋
ex13_3%>%
kbl(caption = "Dataset",escape=F) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam13_3.Group | exam13_3.Y |
|---|---|
| 1 | 2 |
| 1 | 0 |
| 1 | 2 |
| 1 | 3 |
| 1 | 0 |
| 2 | 6 |
| 2 | 4 |
| 2 | 8 |
| 2 | 2 |
| 2 | 4 |
| 3 | 9 |
| 3 | 5 |
| 3 | 6 |
| 3 | 5 |
| 3 | 11 |
| 4 | 2 |
| 4 | 4 |
| 4 | 1 |
| 4 | 0 |
| 4 | 2 |
- 해당 데이터를 사용하여 제곱근변환을 고려해볼 것이다.
- 제곱근변환은 분산이 평균과 같이 증가하는 경향이 있는 경우, 포아송 분포를 따르는 데이터들 혹은 그룹의 분산이 평균들에 비례적일 경우 고려할 수 있다.
Group1 <- subset(ex13_3$exam13_3.Y,ex13_3$exam13_3.Group==1)
Group2 <- subset(ex13_3$exam13_3.Y,ex13_3$exam13_3.Group==2)
Group3 <- subset(ex13_3$exam13_3.Y,ex13_3$exam13_3.Group==3)
Group4 <- subset(ex13_3$exam13_3.Y,ex13_3$exam13_3.Group==4)
ex13_3_split <- data.frame(Group1,Group2,Group3,Group4)
ex13_3_split%>%
kbl(caption = "Dataset of Example 13.3") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Group1 | Group2 | Group3 | Group4 |
|---|---|---|---|
| 2 | 6 | 9 | 2 |
| 0 | 4 | 5 | 4 |
| 2 | 8 | 6 | 1 |
| 3 | 2 | 5 | 0 |
| 0 | 4 | 11 | 2 |
\[\begin{aligned} X^* = \sqrt{X+0.5} \end{aligned}\]제곱근변환
- 위 식을 사용하여 제곱근변환을 시도하겠다.
sqrt_ex13_3 <- sqrt(ex13_3_split+0.5)
sqrt_ex13_3%>%
kbl(caption = "Sqruare root transformation of Example 13.3") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Group1 | Group2 | Group3 | Group4 |
|---|---|---|---|
| 1.5811388 | 2.549510 | 3.082207 | 1.5811388 |
| 0.7071068 | 2.121320 | 2.345208 | 2.1213203 |
| 1.5811388 | 2.915476 | 2.549510 | 1.2247449 |
| 1.8708287 | 1.581139 | 2.345208 | 0.7071068 |
| 0.7071068 | 2.121320 | 3.391165 | 1.5811388 |
parameter <- c("Mean","Variance","Std","Lower CI","Upper CI")
f13_3 <- function(x){
mean <- mean(x)
s2 <- var(x)
s <- sd(x)
L1 <- mean-(qt(0.975,4)*(s/sqrt(length(x))))
L2 <- mean+(qt(0.975,4)*(s/sqrt(length(x))))
res <- list(mean,s2,s,L1,L2)
return(res)
}
Group_1_original <- f13_3(ex13_3_split$Group1)
Group_2_original <- f13_3(ex13_3_split$Group2)
Group_3_original <- f13_3(ex13_3_split$Group3)
Group_4_original <- f13_3(ex13_3_split$Group4)
origin <- data.frame(cbind(parameter,Group_1_original,Group_2_original,Group_3_original,Group_4_original))
origin%>%
kbl(caption = "Result of Original dataset") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| parameter | Group_1\_original | Group_2\_original | Group_3\_original | Group_4\_original |
|---|---|---|---|---|
| Mean | 1.4 | 4.8 | 7.2 | 1.8 |
| Variance | 1.8 | 5.2 | 7.2 | 2.2 |
| Std | 1.341641 | 2.280351 | 2.683282 | 1.48324 |
| Lower CI | -0.2658671 | 1.96857 | 3.868266 | -0.04168533 |
| Upper CI | 3.065867 | 7.63143 | 10.53173 | 3.641685 |
Group_1_sqrtrans <- f13_3(sqrt_ex13_3$Group1)
Group_2_sqrtrans <- f13_3(sqrt_ex13_3$Group2)
Group_3_sqrtrans <- f13_3(sqrt_ex13_3$Group3)
Group_4_sqrtrans <- f13_3(sqrt_ex13_3$Group4)
sqrtrans <- data.frame(cbind(parameter,Group_1_sqrtrans,Group_2_sqrtrans,Group_3_sqrtrans,Group_4_sqrtrans))
sqrtrans%>%
kbl(caption = "Result of logarithmic transformation dataset") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| parameter | Group_1\_sqrtrans | Group_2\_sqrtrans | Group_3\_sqrtrans | Group_4\_sqrtrans |
|---|---|---|---|---|
| Mean | 1.289464 | 2.257753 | 2.74266 | 1.44309 |
| Variance | 0.2966033 | 0.253189 | 0.2222736 | 0.2718643 |
| Std | 0.544613 | 0.5031789 | 0.471459 | 0.5214061 |
| Lower CI | 0.6132377 | 1.632974 | 2.157266 | 0.7956788 |
| Upper CI | 1.96569 | 2.882532 | 3.328053 | 2.090501 |
- 이렇게 변수변환 한 후에는 모수적 검정법을 적용하여 추후 분석을 진행할 수 있다.
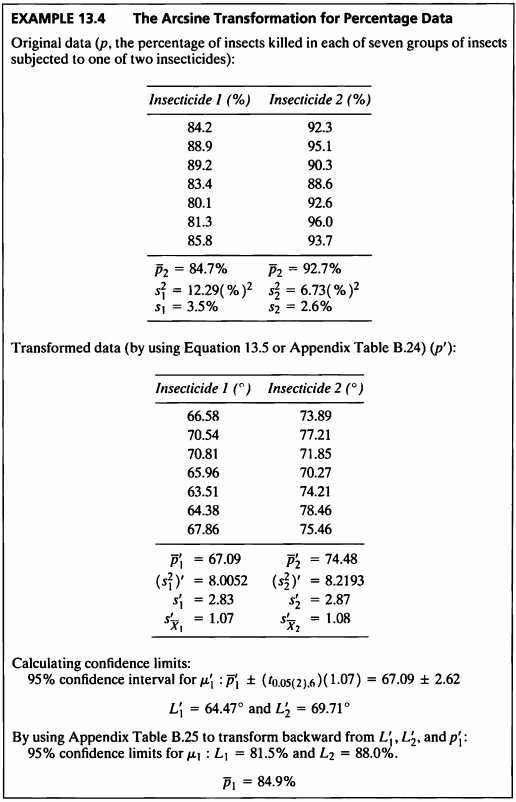
EXAMPLE 13.4
#데이터셋
ex13_4%>%
kbl(caption = "Dataset",escape=F) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam13_4.Group | exam13_4.Y |
|---|---|
| 1 | 84.2 |
| 1 | 88.9 |
| 1 | 89.2 |
| 1 | 83.4 |
| 1 | 80.1 |
| 1 | 81.3 |
| 1 | 85.8 |
| 2 | 92.3 |
| 2 | 95.1 |
| 2 | 90.3 |
| 2 | 88.6 |
| 2 | 92.6 |
| 2 | 96.0 |
| 2 | 93.7 |
- 해당 데이터를 사용하여 역싸인변환을 고려해볼 것이다.
- 역싸인변환은 이항형의 데이터 즉, 0~100% 까지의 퍼센티지 데이터 혹은 0과 1의 비율 데이터에 대해서 고려할 수 있다.
Group1 <- subset(ex13_4$exam13_4.Y,ex13_4$exam13_4.Group==1)
Group2 <- subset(ex13_4$exam13_4.Y,ex13_4$exam13_4.Group==2)
ex13_4_split <- data.frame(Group1,Group2)
ex13_4_split%>%
kbl(caption = "Dataset of Example 13.4") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Group1 | Group2 |
|---|---|
| 84.2 | 92.3 |
| 88.9 | 95.1 |
| 89.2 | 90.3 |
| 83.4 | 88.6 |
| 80.1 | 92.6 |
| 81.3 | 96.0 |
| 85.8 | 93.7 |
\[\begin{aligned} p^* &= arcsin \sqrt{p} \\ &=arcsin\sqrt{\frac{p}{100}} \times \frac{180}{\pi} \end{aligned}\]역싸인변환
- 위 식을 사용하여 역싸인변환을 시도하겠다.
arcsin_ex13_4 <- asin(sqrt(ex13_4_split/100))*(180/pi)
arcsin_ex13_4%>%
kbl(caption = "Arcsine tranformation of Example 13.4") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Group1 | Group2 |
|---|---|
| 66.57851 | 73.88962 |
| 70.53891 | 77.21111 |
| 70.81414 | 71.85347 |
| 65.95645 | 70.26691 |
| 63.50664 | 74.21494 |
| 64.37781 | 78.46304 |
| 67.86261 | 75.46342 |
parameter <- c("Mean","Variance","Std","Lower CI","Upper CI")
f13_4 <- function(x){
mean <- mean(x)
s2 <- var(x)
s <- sd(x)
L1 <- mean-(qt(0.975,6)*(s/sqrt(length(x))))
L2 <- mean+(qt(0.975,6)*(s/sqrt(length(x))))
res <- list(mean,s2,s,L1,L2)
return(res)
}
Group_1_original <- f13_4(ex13_4_split$Group1)
Group_2_original <- f13_4(ex13_4_split$Group2)
origin <- data.frame(cbind(parameter,Group_1_original,Group_2_original))
origin%>%
kbl(caption = "Result of Original dataset") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| parameter | Group_1\_original | Group_2\_original |
|---|---|---|
| Mean | 84.7 | 92.65714 |
| Variance | 12.29333 | 6.729524 |
| Std | 3.506185 | 2.594133 |
| Lower CI | 81.45732 | 90.25797 |
| Upper CI | 87.94268 | 95.05632 |
Group_1_arctrans <- f13_4(arcsin_ex13_4$Group1)
Group_2_arctrans <- f13_4(arcsin_ex13_4$Group2)
arctrans <- data.frame(cbind(parameter,Group_1_arctrans,Group_2_arctrans))
arctrans%>%
kbl(caption = "Result of arcsine transformation dataset") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| parameter | Group_1\_arctrans | Group_2\_arctrans |
|---|---|---|
| Mean | 67.09072 | 74.48036 |
| Variance | 8.017362 | 8.226374 |
| Std | 2.831495 | 2.868166 |
| Lower CI | 64.47203 | 71.82775 |
| Upper CI | 69.70942 | 77.13297 |
SAS 프로그램 결과
SAS 접기/펼치기 버튼
13장
LIBNAME ex 'C:\Biostat';
RUN;
/*13장 연습문제 불러오기*/
%macro chap13(name=,no=);
%do i=1 %to &no.;
PROC IMPORT DBMS=excel
DATAFILE="C:\Biostat\data_chap13"
OUT=ex.&name.&i. REPLACE;
RANGE="exam13_&i.$";
RUN;
%end;
%mend;
%chap13(name=ex13_,no=4);
EXAMPLE 13.1
DATA ex_13_1_a;
SET ex.ex13_1;
WHERE part = "a)";
RUN;
DATA ex_13_1_b;
SET ex.ex13_1;
WHERE part = "b)";
RUN;
DATA ex_13_1_c;
SET ex.ex13_1;
WHERE part = "c)";
RUN;
title "Example13.1";
PROC GLM DATA=ex_13_1_a;
CLASS a b;
MODEL mean = a b a*b;
RUN;
PROC GLM DATA=ex_13_1_b;
CLASS a b;
MODEL mean = a b a*b;
RUN;
PROC GLM DATA=ex_13_1_c;
CLASS a b;
MODEL mean = a b a*b;
RUN;
The GLM Procedure
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| A | 3 | 1 2 3 |
| B | 2 | 1 2 |
| Number of Observations Read | 6 |
|---|---|
| Number of Observations Used | 6 |
The GLM Procedure
Dependent Variable: mean mean
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Model | 5 | 383.3333333 | 76.6666667 | . | . |
| Error | 0 | 0.0000000 | . | ||
| Corrected Total | 5 | 383.3333333 |
| R-Square | Coeff Var | Root MSE | mean Mean |
|---|---|---|---|
| 1.000000 | . | . | 23.33333 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| A | 2 | 233.3333333 | 116.6666667 | . | . |
| B | 1 | 150.0000000 | 150.0000000 | . | . |
| A*B | 2 | 0.0000000 | 0.0000000 | . | . |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| A | 2 | 233.3333333 | 116.6666667 | . | . |
| B | 1 | 150.0000000 | 150.0000000 | . | . |
| A*B | 2 | 0.0000000 | 0.0000000 | . | . |
The GLM Procedure
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| A | 3 | 1 2 3 |
| B | 2 | 1 2 |
| Number of Observations Read | 6 |
|---|---|
| Number of Observations Used | 6 |
The GLM Procedure
Dependent Variable: mean mean
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Model | 5 | 8000.000000 | 1600.000000 | . | . |
| Error | 0 | 0.000000 | . | ||
| Corrected Total | 5 | 8000.000000 |
| R-Square | Coeff Var | Root MSE | mean Mean |
|---|---|---|---|
| 1.000000 | . | . | 50.00000 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| A | 2 | 5700.000000 | 2850.000000 | . | . |
| B | 1 | 1666.666667 | 1666.666667 | . | . |
| A*B | 2 | 633.333333 | 316.666667 | . | . |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| A | 2 | 5700.000000 | 2850.000000 | . | . |
| B | 1 | 1666.666667 | 1666.666667 | . | . |
| A*B | 2 | 633.333333 | 316.666667 | . | . |
The GLM Procedure
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| A | 3 | 1 2 3 |
| B | 2 | 1 2 |
| Number of Observations Read | 6 |
|---|---|
| Number of Observations Used | 6 |
The GLM Procedure
Dependent Variable: mean mean
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Model | 5 | 0.75420000 | 0.15084000 | . | . |
| Error | 0 | 0.00000000 | . | ||
| Corrected Total | 5 | 0.75420000 |
| R-Square | Coeff Var | Root MSE | mean Mean |
|---|---|---|---|
| 1.000000 | . | . | 1.570000 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| A | 2 | 0.61920000 | 0.30960000 | . | . |
| B | 1 | 0.13500000 | 0.13500000 | . | . |
| A*B | 2 | 0.00000000 | 0.00000000 | . | . |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| A | 2 | 0.61920000 | 0.30960000 | . | . |
| B | 1 | 0.13500000 | 0.13500000 | . | . |
| A*B | 2 | 0.00000000 | 0.00000000 | . | . |
EXAMPLE 13.2
title "Example13.2";
PROC MEANS DATA=ex.ex13_2 MEAN STD VAR;
CLASS group;
VAR Y;
RUN;
MEANS 프로시저
| 분석 변수: Y Y | ||||
|---|---|---|---|---|
| Group | 관측값 수 | 평균 | 표준편차 | 분산 |
| 1 | 5 | 3.2800000 | 0.2863564 | 0.0820000 |
| 2 | 5 | 6.9400000 | 0.6024948 | 0.3630000 |
DATA ex13_2_log;
SET ex.ex13_2;
log_y=round(log10(y+1), 0.00001);
RUN;
PROC MEANS DATA=ex13_2_log MEAN STD VAR;
CLASS group;
VAR log_y;
RUN;
MEANS 프로시저
| 분석 변수: log_y | ||||
|---|---|---|---|---|
| Group | 관측값 수 | 평균 | 표준편차 | 분산 |
| 1 | 5 | 0.6306560 | 0.0293030 | 0.000858666 |
| 2 | 5 | 0.8988200 | 0.0329646 | 0.0010867 |
DATA log_g1;
SET ex13_2_log;
WHERE group = 1;
RUN;
DATA log_g2;
SET ex13_2_log;
WHERE group = 2;
RUN;
PROC IML;
USE log_g1;
READ all;
CLOSE log_g1;
t=round(abs(quantile("t",0.05/2, 4)),0.001);
L1_log = round(mean(log_y) -(t*0.01310), 0.00001);
L2_log = round(mean(log_y) +(t*0.01310), 0.00001);
L1_origin=round((10**L1_log)-1, 0.01);
L2_origin=round((10**L2_log)-1, 0.01);
print L1_log L2_log L1_origin L2_origin;
RUN;
QUIT;
| L1_log | L2_log | L1_origin | L2_origin |
|---|---|---|---|
| 0.59429 | 0.66702 | 2.93 | 3.65 |
PROC IML;
USE log_g2;
READ all;
CLOSE log_g2;
t=round(abs(quantile("t",0.05/2, 4)),0.001);
L1_log = round(mean(log_y) -(t*0.01310), 0.00001);
L2_log = round(mean(log_y) +(t*0.01310), 0.00001);
L1_origin=round((10**L1_log)-1, 0.01);
L2_origin=round((10**L2_log)-1, 0.01);
print L1_log L2_log L1_origin L2_origin;
RUN;
QUIT;
| L1_log | L2_log | L1_origin | L2_origin |
|---|---|---|---|
| 0.86245 | 0.93519 | 6.29 | 7.61 |
EXAMPLE 13.3
PROC TABULATE DATA=ex.ex13_3;
CLASS Group;
VAR Y;
TABLE Y*(MEAN*F=6.1 VAR*F=6.1), Group;
RUN;
| Group | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Y | Mean | 1.4 | 4.8 | 7.2 | 1.8 |
| Var | 1.8 | 5.2 | 7.2 | 2.2 | |
DATA ex13_3_eq13_2;
SET ex.ex13_3;
transY = sqrt(Y+0.5);
RUN;
PROC TABULATE DATA=ex13_3_eq13_2 out=ex13_3_out;
CLASS Group;
VAR transY;
TABLE transY*(MEAN*F=6.3 VAR*F=6.3 lclm*F=6.3 uclm*F=6.3), Group;
RUN;
| Group | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| transY | Mean | 1.289 | 2.258 | 2.743 | 1.443 |
| Var | 0.297 | 0.253 | 0.222 | 0.272 | |
| 95_LCLM | 0.613 | 1.633 | 2.157 | 0.796 | |
| 95_UCLM | 1.966 | 2.883 | 3.328 | 2.091 | |
DATA ex13_3_original_unit;
SET ex13_3_out;
original_unit_mean = round( (transY_Mean)**2 - 0.5, 0.1);
original_unit_LCLM = round( (transY_LCLM)**2 - 0.5, 0.1);
original_unit_UCLM = round( (transY_UCLM)**2 - 0.5, 0.1);
RUN;
PROC PRINT DATA=ex13_3_original_unit;
VAR Group original_unit_mean original_unit_LCLM original_unit_UCLM;
RUN;
| OBS | Group | original_unit_mean | original_unit_LCLM | original_unit_UCLM |
|---|---|---|---|---|
| 1 | 1 | 1.2 | -0.1 | 3.4 |
| 2 | 2 | 4.6 | 2.2 | 7.8 |
| 3 | 3 | 7.0 | 4.2 | 10.6 |
| 4 | 4 | 1.6 | 0.1 | 3.9 |
EXAMPLE 13.4
title "Example13.4";
PROC TABULATE DATA=ex.ex13_4;
CLASS Group;
VAR Y;
TABLE Y*(MEAN*F=6.1 VAR*F=6.2 STD*F=6.1 lclm*F=6.2 uclm*F=6.2), Group;
RUN;
| Group | |||
|---|---|---|---|
| 1 | 2 | ||
| Y | Mean | 84.7 | 92.7 |
| Var | 12.29 | 6.73 | |
| Std | 3.5 | 2.6 | |
| 95_LCLM | 81.46 | 90.26 | |
| 95_UCLM | 87.94 | 95.06 | |
DATA exam13_4_trans;
SET ex.ex13_4;
pi=constant("PI") ;
transY=arsin(sqrt(Y/100))*(180/pi) ;
RUN;
PROC TABULATE DATA=exam13_4_trans;
CLASS Group;
VAR transY;
TABLE transY*(MEAN*F=6.2 VAR*F=6.4 STD*F=6.2 lclm*F=6.2 uclm*F=6.2), Group;
RUN;
| Group | |||
|---|---|---|---|
| 1 | 2 | ||
| transY | Mean | 67.09 | 74.48 |
| Var | 8.0174 | 8.2264 | |
| Std | 2.83 | 2.87 | |
| 95_LCLM | 64.47 | 71.83 | |
| 95_UCLM | 69.71 | 77.13 | |
교재: Biostatistical Analysis (5th Edition) by Jerrold H. Zar
**이 글은 22학년도 1학기 의학통계방법론 과제 자료들을 정리한 글 입니다.**
