[의학통계방법론] Ch12. Two-Factor Analysis of Variance
Two-Factor Analysis of Variance
R 프로그램 결과
R 접기/펼치기 버튼
패키지 설치된 패키지 접기/펼치기 버튼
getwd()
## [1] "C:/Biostat"
library("readxl")
library("dplyr")
library("kableExtra")
library("broom")
library("ggplot2")
library("agricolae")
library("nonpar")
엑셀파일불러오기
#모든 시트를 하나의 리스트로 불러오는 함수
read_excel_allsheets <- function(file, tibble = FALSE) {
sheets <- readxl::excel_sheets(file)
x <- lapply(sheets, function(X) readxl::read_excel(file, sheet = X))
if(!tibble) x <- lapply(x, as.data.frame)
names(x) <- sheets
x
}
12장
12장 연습문제 불러오기
#data_chap12에 연습문제 12장 모든 문제 저장
data_chap12 <- read_excel_allsheets("data_chap12.xls")
#연습문제 각각 데이터 생성
for (x in 1:length(data_chap12)){
assign(paste0('ex12_',c(1,4,5,6))[x],data_chap12[x])
}
#연습문제 데이터 형식을 리스트에서 데이터프레임으로 변환
for (x in 1:length(data_chap12)){
assign(paste0('ex12_',c(1,4,5,6))[x],data.frame(data_chap12[x]))
}
EXAMPLE 12.1
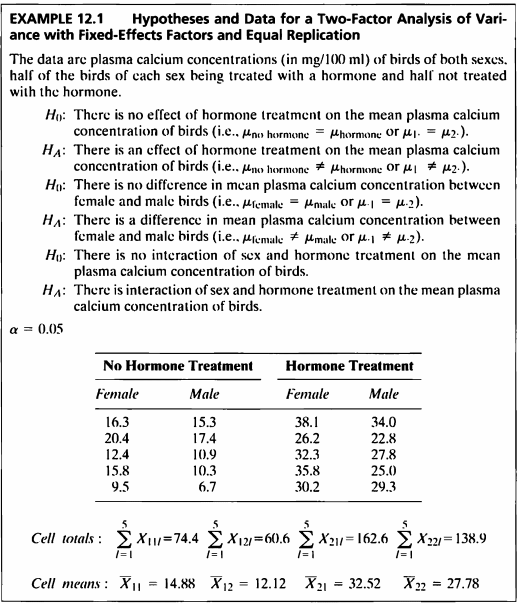
#데이터셋
ex12_1%>%
kbl(caption = "Dataset",escape=F) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam12_1.No | exam12_1.Hormone | exam12_1.Sex | exam12_1.plasma_calcium |
|---|---|---|---|
| 1 | No Hormone | Female | 16.3 |
| 2 | No Hormone | Female | 20.4 |
| 3 | No Hormone | Female | 12.4 |
| 4 | No Hormone | Female | 15.8 |
| 5 | No Hormone | Female | 9.5 |
| 6 | No Hormone | Male | 15.3 |
| 7 | No Hormone | Male | 17.4 |
| 8 | No Hormone | Male | 10.9 |
| 9 | No Hormone | Male | 10.3 |
| 10 | No Hormone | Male | 6.7 |
| 11 | Yes Hormone | Female | 38.1 |
| 12 | Yes Hormone | Female | 26.2 |
| 13 | Yes Hormone | Female | 32.3 |
| 14 | Yes Hormone | Female | 35.8 |
| 15 | Yes Hormone | Female | 30.2 |
| 16 | Yes Hormone | Male | 34.0 |
| 17 | Yes Hormone | Male | 22.8 |
| 18 | Yes Hormone | Male | 27.8 |
| 19 | Yes Hormone | Male | 25.0 |
| 20 | Yes Hormone | Male | 29.3 |
- 해당 데이터는 성별에 따른 새의 혈청 내 칼슘 농도를 기록한 데이터이다.
- 호르몬 치료 유무와 성별에 따른 혈청 내 칼슘 농도에 차이가 있는지 이요인 분산분석을 시행한다.
nf <- subset(ex12_1$exam12_1.plasma_calcium, ex12_1$exam12_1.Hormone=="No Hormone" & ex12_1$exam12_1.Sex=="Female")
nm <- subset(ex12_1$exam12_1.plasma_calcium, ex12_1$exam12_1.Hormone=="No Hormone" & ex12_1$exam12_1.Sex=="Male")
yf <- subset(ex12_1$exam12_1.plasma_calcium, ex12_1$exam12_1.Hormone=="Yes Hormone" & ex12_1$exam12_1.Sex=="Female")
ym <- subset(ex12_1$exam12_1.plasma_calcium, ex12_1$exam12_1.Hormone=="Yes Hormone" & ex12_1$exam12_1.Sex=="Male")
ex12_1a <- data.frame(no_hormone_female=nf,no_hormone_male=nm,yes_hormone_female=yf,yes_hormone_male=ym)
ex12_1a%>%
kbl(caption = "Dataset of ex12_1") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| no_hormone_female | no_hormone_male | yes_hormone_female | yes_hormone_male |
|---|---|---|---|
| 16.3 | 15.3 | 38.1 | 34.0 |
| 20.4 | 17.4 | 26.2 | 22.8 |
| 12.4 | 10.9 | 32.3 | 27.8 |
| 15.8 | 10.3 | 35.8 | 25.0 |
| 9.5 | 6.7 | 30.2 | 29.3 |
cell_totals <- c(sum(nf),sum(nm),sum(yf),sum(ym))
cell_means <- c(mean(nf),mean(nm),mean(yf),mean(ym))
cell <- data.frame(rbind(cell_totals,cell_means))
names(cell) <- c("X_11","X_12","X_21","X_22")
cell%>%
kbl(caption = "Cell totals and Cell means") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| X_11 | X_12 | X_21 | X_22 | |
|---|---|---|---|---|
| cell_totals | 74.40 | 60.60 | 162.60 | 138.90 |
| cell_means | 14.88 | 12.12 | 32.52 | 27.78 |
EXAMPLE 12.1a
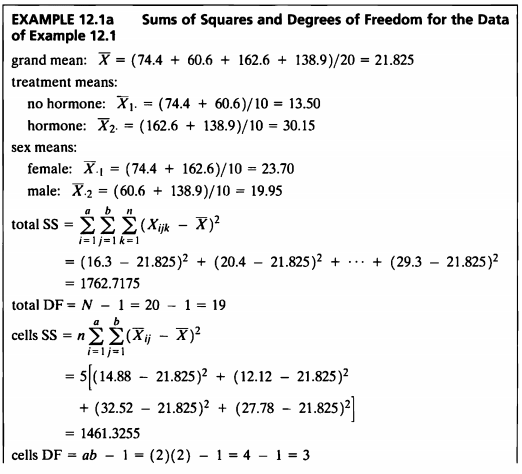
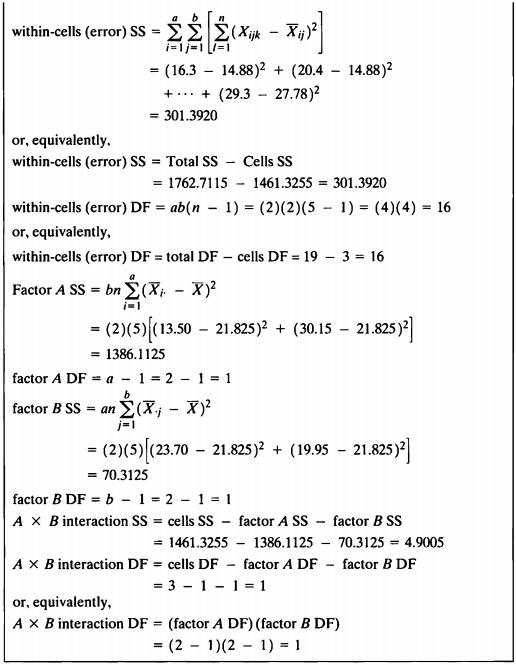
- 예제에서 나온 식으로 제곱합 및 자유도를 구하는 함수를 직접 작성하여 ANOVA table을 작성하여 보았다.
f12_1a <- function(x){
N <- (length(x)*nrow(x))
grand_mean <- sum(x)/N
no_hormone_means <- sum(x[,1:2])/10
hormone_means <- sum(x[,3:4])/10
female_means <- sum(x[,c(1,3)])/10
male_means <- sum(x[,c(2,4)])/10
total_SS <- sum((x-grand_mean)^2)
total_DF <- N-1
cells <- vector()
for (i in 1:4){
cells[i] <- (mean(ex12_1a[,i]) - grand_mean)^2
}
cells_SS <- sum(5*cells)
cells_DF <- 3
within_cells_SS <- total_SS - cells_SS
within_cells_DF <- total_DF - cells_DF
Factor_A_SS <- 2*5*((no_hormone_means-grand_mean)^2+(hormone_means-grand_mean)^2)
Factor_A_DF <- 1
Factor_B_SS <- 2*5*((female_means-grand_mean)^2+(male_means-grand_mean)^2)
Factor_B_DF <- 1
AXB_interaction_SS <- cells_SS-Factor_A_SS-Factor_B_SS
AXB_interaction_DF <- cells_DF - Factor_A_DF - Factor_B_DF
SS <- c(total_SS,cells_SS,Factor_A_SS,Factor_B_SS,round(AXB_interaction_SS,4),round(within_cells_SS,4))
DF <- c(total_DF,cells_DF,Factor_A_DF,Factor_B_DF,AXB_interaction_DF,within_cells_DF)
source <- c("Total","Cells","Factor A (hormone)","Facor B (sex)","A X B","Within-Cells (Error)")
MS <- c("","",Factor_A_SS/Factor_A_DF,Factor_B_SS/Factor_B_DF,round(AXB_interaction_SS/AXB_interaction_DF,4),within_cells_SS/within_cells_DF)
res <- data.frame(source,SS,DF,MS)
return(res)
}
f12_1a(ex12_1a)%>%
kbl(caption = "Analysis of Variance Summary table") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| source | SS | DF | MS |
|---|---|---|---|
| Total | 1762.7175 | 19 | |
| Cells | 1461.3255 | 3 | |
| Factor A (hormone) | 1386.1125 | 1 | 1386.1125 |
| Facor B (sex) | 70.3125 | 1 | 70.3125 |
| A X B | 4.9005 | 1 | 4.9005 |
| Within-Cells (Error) | 301.3920 | 16 | 18.837 |
EXAMPLE 12.2
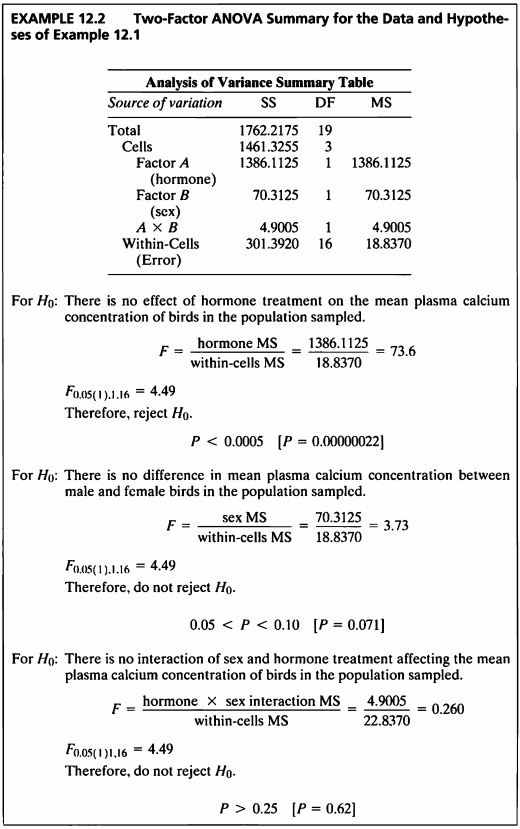
ex12_1$Hormone <- as.factor(ex12_1$exam12_1.Hormone)
ex12_1$Sex <- as.factor(ex12_1$exam12_1.Sex)
- 이원분산분석을 진행하기 전에 정규성과 등분산성, 독립성을 만족하는지 부터 확인하도록 한다.
Shapiro-Wilk Test로 그룹별 정규성을 평가하였다.
ex12_1 %>% group_by(exam12_1.Hormone, exam12_1.Sex) %>%
summarise(shapiro_test = shapiro.test(exam12_1.plasma_calcium)$p.value)
## # A tibble: 4 × 3
## # Groups: exam12_1.Hormone [2]
## exam12_1.Hormone exam12_1.Sex shapiro_test
## <chr> <chr> <dbl>
## 1 No Hormone Female 0.931
## 2 No Hormone Male 0.800
## 3 Yes Hormone Female 0.949
## 4 Yes Hormone Male 0.926
- 4그룹 모두 p-value가 0.05보다 매우 크므로 모집단이 정규성을 따르지 않는다는 대립가설을 채택할 충분한 근거가 없다.
- 따라서 정규성을 만족한다는 사실을 바탕으로 등분산성 검정을 진행한다.
Bartlett’s Test를 통해 등분산성 검정을 진행하였다.
bartlett.test(ex12_1$exam12_1.plasma_calcium~ex12_1$exam12_1.Hormone)
##
## Bartlett test of homogeneity of variances
##
## data: ex12_1$exam12_1.plasma_calcium by ex12_1$exam12_1.Hormone
## Bartlett's K-squared = 0.19987, df = 1, p-value = 0.6548
bartlett.test(ex12_1$exam12_1.plasma_calcium~ex12_1$exam12_1.Sex)
##
## Bartlett test of homogeneity of variances
##
## data: ex12_1$exam12_1.plasma_calcium by ex12_1$exam12_1.Sex
## Bartlett's K-squared = 0.091482, df = 1, p-value = 0.7623
- p-value가 모두 0.05보다 크므로 등분산성이 아니라는 대립가설을 채택할 근거가 없다.
\[\begin{aligned} H_0 &: There\ is\ no\ effect\ of\ hormone\ treatment\ on\ the\ mean\ plasma\ calcium\ concentration\ of\ birds\ \\ H_A &: There\ is\ an\ effect\ of\ hormone\ treatment\ on\ the\ mean\ plasma\ calcium\ concentration\ of\ birds\ \\ \end{aligned}\]첫번째 가설
f12_2_1 <- function(x){
f <- f12_1a(x)$SS[3]/as.numeric(f12_1a(x)$MS[6])
f_0.05 <- qf(0.95,1,16)
p_value <- 1-pf(f,1,16)
if (f<f_0.05){
reject_H0 <- "No"
}else{
reject_H0 <- "Yes"
}
res <- data.frame(f,f_0.05,p_value,reject_H0)
return(res)
}
f12_2_1(ex12_1a)%>%
kbl(caption = "Result of hypotheses for Hormone treatment") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| f | f_0.05 | p_value | reject_H0 |
|---|---|---|---|
| 73.58457 | 4.493999 | 2e-07 | Yes |
- F-값이 73.6으로 임계값 4.49보다 큰 값이 나왔고, p-value가 0.05보다 매우 작으므로 유의수준 5%하에 귀무가설을 기각한다.
- 따라서 호르몬 치료에 따른 혈청 내 칼슘 함량의 평균이 다르다고 할 수 있다.
\[\begin{aligned} H_0 &: There\ is\ no\ difference\ \ in\ mean\ plasma\ calcium\ concentration\ between\ female\ and\ male\ birds\ \\ H_A &: There\ is\ a\ difference\ \ in\ mean\ plasma\ calcium\ concentration\ between\ female\ and\ male\ birds\ \\ \end{aligned}\]두번째 가설
f12_2_2 <- function(x){
f <- f12_1a(x)$SS[4]/as.numeric(f12_1a(x)$MS[6])
f_0.05 <- qf(0.95,1,16)
p_value <- 1-pf(f,1,16)
if (f<f_0.05){
reject_H0 <- "No"
}else{
reject_H0 <- "Yes"
}
res <- data.frame(f,f_0.05,p_value,reject_H0)
return(res)
}
f12_2_2(ex12_1a)%>%
kbl(caption = "Result of hypotheses for sex") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| f | f_0.05 | p_value | reject_H0 |
|---|---|---|---|
| 3.73268 | 4.493999 | 0.0712638 | No |
- F-값이 3.73으로 임계값 4.49보다 작은 값이 나왔고, p-value가 0.05보다 크므로 유의수준 5%하에 귀무가설을 기각할 수 없다.
- 따라서 성별에 따른 혈청 내 칼슘 함량의 평균이 다르다고 할 수 없다.
\[\begin{aligned} H_0 &: There\ is\ no\ interaction\ of\ sex\ and\ hormone\ treatment\ on\ the\ mean\ plasma\ calcium\ concentration\ of\ birds\ \\ H_A &: There\ is\ interaction\ of\ sex\ and\ hormone\ treatment\ on\ the\ mean\ plasma\ calcium\ concentration\ of\ birds\ \\ \end{aligned}\]세번째 가설
f12_2_3 <- function(x){
f <- f12_1a(x)$SS[5]/as.numeric(f12_1a(x)$MS[6])
f_0.05 <- qf(0.95,1,16)
p_value <- 1-pf(f,1,16)
if (f<f_0.05){
reject_H0 <- "No"
}else{
reject_H0 <- "Yes"
}
res <- data.frame(f,f_0.05,p_value,reject_H0)
return(res)
}
f12_2_3(ex12_1a)%>%
kbl(caption = "Result of hypotheses for interaction") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| f | f_0.05 | p_value | reject_H0 |
|---|---|---|---|
| 0.2601529 | 4.493999 | 0.6169788 | No |
- F-값이 0.26으로 임계값 4.49보다 작은 값이 나왔고, p-value가 0.05보다 크므로 유의수준 5%하에 귀무가설을 기각할 수 없다.
- 따라서 호르몬과 성별의 교호작용 효과가 있다고 할 수 없다.
aov()를 사용하여 이원분산분석을 실시한다.
library(broom)
aov(exam12_1.plasma_calcium~exam12_1.Hormone*exam12_1.Sex, data=ex12_1) %>%
tidy() %>%
rename(" "="term","Sum Sq"="sumsq","Mean Sq"="meansq","F value"="statistic","Pr(>F)"="p.value") %>%
kable(caption = "Two-factor ANOVA",booktabs = TRUE, valign = 't')%>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| df | Sum Sq | Mean Sq | F value | Pr(\>F) | |
|---|---|---|---|---|---|
| exam12_1.Hormone | 1 | 1386.1125 | 1386.1125 | 73.5845676 | 0.0000002 |
| exam12_1.Sex | 1 | 70.3125 | 70.3125 | 3.7326804 | 0.0712638 |
| exam12_1.Hormone:exam12_1.Sex | 1 | 4.9005 | 4.9005 | 0.2601529 | 0.6169788 |
| Residuals | 16 | 301.3920 | 18.8370 | NA | NA |
stripchart() 함수로 1차원 산점도를 확인하여 본다.
par(mfrow=c(1,2))
stripchart(ex12_1$exam12_1.plasma_calcium~ex12_1$exam12_1.Hormone , vertical=T, xlab="Hormone", ylab="calcium", method="jitter", col="#8dd3c7", pch=16)
stripchart(ex12_1$exam12_1.plasma_calcium~ex12_1$exam12_1.Sex, vertical=T, xlab="Female / male", ylab="calcium", method="jitter", col="#007266", pch=16)
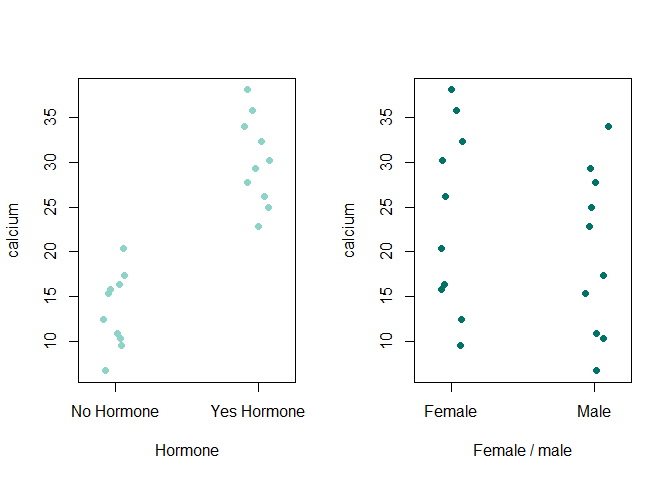
- stripchart() 함수는 sample size가 작을 때 box plot을 대신하기 좋다.
interaction.plot(ex12_1$exam12_1.Hormone,ex12_1$exam12_1.Sex,ex12_1$exam12_1.plasma_calcium, col=c("orange","#8f7450"), bty="l", main="interaction plot", xlab="Hormone", ylab="mean of calcium", trace.label = "Sex")
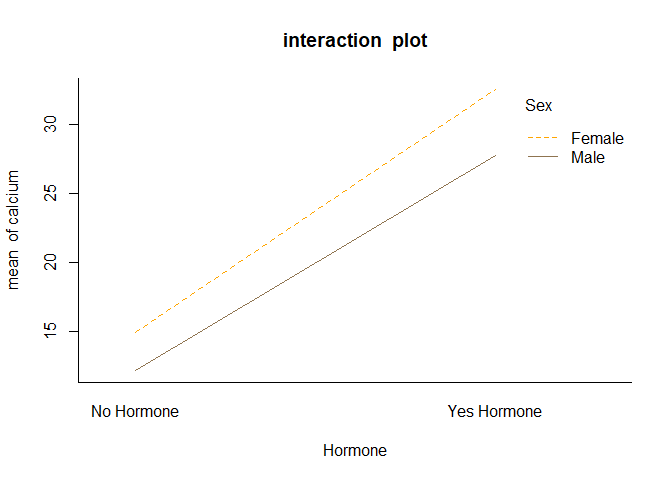
- 교호작용 그래프를 보면 서로 겹치는 부분이 없어 성별과 호르몬 간 교호작용이 존재하지 않음을 확인할 수 있으며, 성별이 여자이고 호르몬치료를 받았을 때 혈청 내 칼슘의 농도가 최대가 됨을 알 수 있다.
EXAMPLE 12.2a
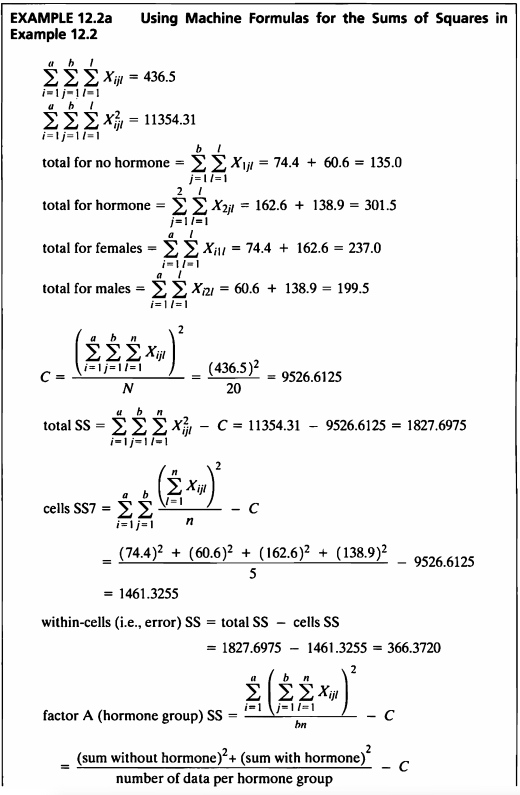
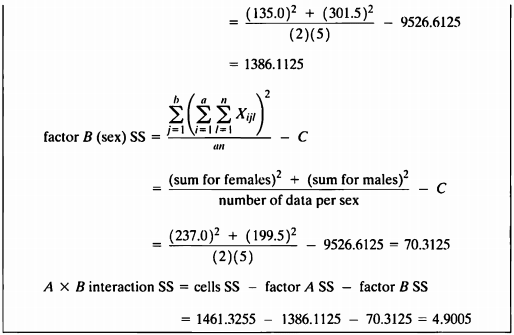
“Machine Formulas”라는 사용하여 Example 12.1a에서 구한 결과와 같은 값을 구하여 보자.
f12_2a <- function(x){
N <- (length(x)*nrow(x))
Xi <- sum(x)
Xi2 <- sum(x^2)
C <- sum(x)^2/N
no_hormone <- sum(x[,1:2])
hormone <- sum(x[,3:4])
females <- sum(x[,c(1,3)])
males <- sum(x[,c(2,4)])
total_SS <- Xi2-C
aa <- vector()
for (i in 1:4){
aa[i] <- sum(ex12_1a[i])^2
}
cells_SS7 <- (sum(aa)/5) -C
within_cells_SS <- total_SS-cells_SS7
Factor_A_SS <- ((no_hormone^2+hormone^2)/10)-C
Factor_B_SS <- ((females^2+males^2)/10)-C
AXB_interaction_SS <- cells_SS7-Factor_A_SS-Factor_B_SS
res <- data.frame(total_SS,cells_SS7,within_cells_SS,Factor_A_SS,Factor_B_SS,AXB_interaction_SS)
return(res)
}
f12_2a(ex12_1a)%>%
kbl(caption = "Result of Machine formula") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| total_SS | cells_SS7 | within_cells_SS | Factor_A\_SS | Factor_B\_SS | AXB_interaction_SS |
|---|---|---|---|---|---|
| 1762.717 | 1461.325 | 301.392 | 1386.112 | 70.3125 | 4.9005 |
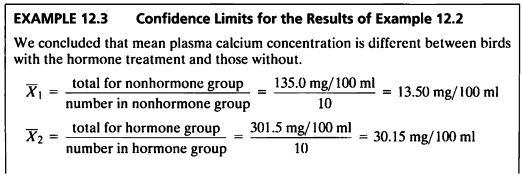
EXAMPLE 12.3

Xbar_1 = mean(ex12_1[ex12_1$exam12_1.Hormone=="No Hormone",]$exam12_1.plasma_calcium)
Xbar_2 = mean(ex12_1[ex12_1$exam12_1.Hormone=="Yes Hormone",]$exam12_1.plasma_calcium)
No.Hormone = ex12_1[ex12_1$exam12_1.Hormone=="No Hormone",]$exam12_1.plasma_calcium
Yes.Hormone = ex12_1[ex12_1$exam12_1.Hormone=="Yes Hormone",]$exam12_1.plasma_calcium
Female=ex12_1[ex12_1$exam12_1.Sex=="Female",]$exam12_1.plasma_calcium
Male=ex12_1[ex12_1$exam12_1.Sex=="Male",]$exam12_1.plasma_calcium
Xbar_pooled = (sum(Female)+sum(Male))/(length(Female)+length(Male))
- 호르몬을 처리하지 않은 새들의 plasma calcium 농도의 표본평균은 13.50mg/100ml이고,
- 호르몬을 처리한 새들의 plasma calcium 농도의 표본평균은 30.15 mg/100ml이었다.
CI_ex12 <- function(x){
exam12_3_results = vector()
sample.mean <- mean(x)
exam12_3_results <- c(exam12_3_results, "mean"=sample.mean)
sample.s2 <- 18.8370
sample.n <- length(x)
t.score = round(qt(p=0.05/2, df=16,lower.tail=F),3)
margin.error <- t.score * sqrt(sample.s2/sample.n)
lower.bound <- round(sample.mean - margin.error, 2)
upper.bound <- round(sample.mean + margin.error, 2)
CI = paste(lower.bound, ",", upper.bound)
exam12_3_results <- c(exam12_3_results, "95% CI"=CI)
exam12_3_results <- as.data.frame(exam12_3_results)
return(exam12_3_results)
}
CI_ex12(No.Hormone)
## exam12_3_results
## mean 13.5
## 95% CI 10.59 , 16.41
CI_ex12(Yes.Hormone)
## exam12_3_results
## mean 30.15
## 95% CI 27.24 , 33.06
CI_ex12(Female)
## exam12_3_results
## mean 23.7
## 95% CI 20.79 , 26.61
CI_ex12(Male)
## exam12_3_results
## mean 19.95
## 95% CI 17.04 , 22.86
- 호르몬을 처리하지 않은 새들의 plasma calcium 농도의 모평균에 대한 95% 신뢰구간 (10.59mg/100ml , 16.41mg/100ml)
- 호르몬 처리를 한 새들의 plasma calcium 농도의 모평균에 대한 95% 신뢰구간 (27.24mg/100ml, 33.06mg/100ml)
- 암컷 새들의 plasma calcium 농도의 모평균에 대한 95% 신뢰구간 (20.79mg/100ml, 26.61mg/100ml)
- 수컷 새들의 plasma calcium 농도의 모평균에 대한 95% 신뢰구간 (17.04mg/100ml, 22.86mg/100ml)
CI_ex12_difference <- function(x){
exam12_3_results = vector()
sample.mean <- mean(x)
exam12_3_results <- c(exam12_3_results, "mean"=sample.mean)
sample.s2 <- 18.8370
sample.n <- length(x)
q.score = round(qtukey(p=0.95,2,16),3)
margin.error <- q.score * sqrt(sample.s2/sample.n)
lower.bound <- round(sample.mean - margin.error, 2)
upper.bound <- round(sample.mean + margin.error, 2)
CI = paste(lower.bound, ",", upper.bound)
exam12_3_results <- c(exam12_3_results, "95% CI"=CI)
exam12_3_results <- as.data.frame(exam12_3_results)
return(exam12_3_results)
}
CI_ex12_difference(No.Hormone - Yes.Hormone)
## exam12_3_results
## mean -16.65
## 95% CI -20.76 , -12.54
CI_ex12_difference(Female - Male)
## exam12_3_results
## mean 3.75
## 95% CI -0.36 , 7.86
- 호르몬을 처리하지 않은 새들의 plasma calcium 농도의 모평균과 호르몬 처리를 한 새들의 plasma calcium 농도의 모평균의 차에 대한 95% 신뢰구간 (-20.76mg/100ml, -12.54mg/100ml)
- 신뢰구간이 0을 포함하지 않으므로 유의수준 5%하에 호르몬 처리에 따라 새들의 plasma calcium 농도의 모평균에는 차이가 있다고 볼 수 있다.
- 암컷 새들의 plasma calcium 농도의 모평균과 수컷 새들의 plasma calcium 농도의 모평균의 차에 대한 95% 신뢰구간 (-0.36mg/100ml, 7.86mg/100ml)
- 신뢰구간이 0을 포함하므로, 유의수준 5%하에 plasma calcium 농도의 모평균은 암컷과 수컷 새들 간에 유의미한 차이가 없다고 말할 수 있다.
CI_ex12_pooled <- function(x){
exam12_3_pooled = vector()
exam12_3_pooled = c(exam12_3_pooled, "Xbar_pooed"=x)
t.score = qt(p=0.05/2, df=16,lower.tail=F)
s2=18.8370
margin = t.score * sqrt(s2/(length(Female)+length(Male)))
lower.bound = round(x - margin, 3)
upper.bound = round(x + margin, 3)
CI = paste(lower.bound, ",", upper.bound)
exam12_3_results <- c(exam12_3_pooled, "95% CI"=CI)
exam12_3_results <- as.data.frame(exam12_3_results)
return(exam12_3_results)
}
CI_ex12_pooled(Xbar_pooled)
## exam12_3_results
## Xbar_pooed 21.825
## 95% CI 19.768 , 23.882
- pooled population mean에 대한 95% 신뢰구간은 (19.76mg/100ml, 23.88mg/100ml)이다.
EXAMPLE 12.4
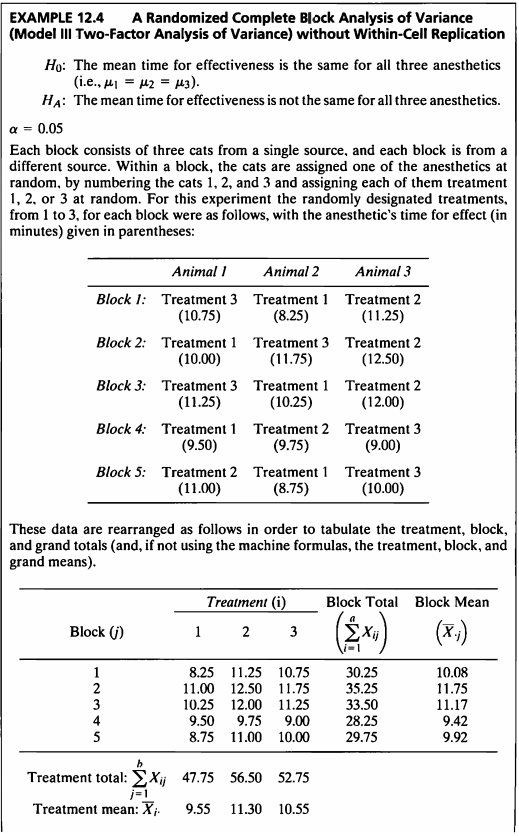
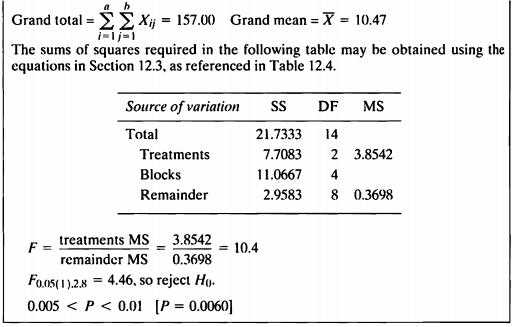
- 해당 데이터는 5개의 블록으로 구성되어 있으며, 한 블록에 세 마리의 고양이로 구성되어있다.
한 블록 안에서 고양이들은 세가지 마취제 중 하나를 무작위로 배정받는다.
배정받은 마취제의 효과 시간(분)을 기록한 데이터이다. - 블록 내에 마취제를 투여함에 있어서는 반복을 하지 않으며, 고양이들이 랜덤으로 설정되었으며 이러한 반복이 없는 경우를 난괴법(Randomized Block Design)이라 한다.
#데이터셋
ex12_4%>%
kbl(caption = "Dataset",escape=F) %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam12_4.Treatment | exam12_4.Block | exam12_4.Time |
|---|---|---|
| 1 | 1 | 8.25 |
| 1 | 2 | 11.00 |
| 1 | 3 | 10.25 |
| 1 | 4 | 9.50 |
| 1 | 5 | 8.75 |
| 2 | 1 | 11.25 |
| 2 | 2 | 12.50 |
| 2 | 3 | 12.00 |
| 2 | 4 | 9.75 |
| 2 | 5 | 11.00 |
| 3 | 1 | 10.75 |
| 3 | 2 | 11.75 |
| 3 | 3 | 11.25 |
| 3 | 4 | 9.00 |
| 3 | 5 | 10.00 |
library(ggplot2)
ex12_4$exam12_4.Treatment <- as.factor(ex12_4$exam12_4.Treatment)
ex12_4$exam12_4.Block <- as.factor(ex12_4$exam12_4.Block)
ggplot(ex12_4, aes(x = exam12_4.Block, y = exam12_4.Time, fill=exam12_4.Block)) +
geom_boxplot()+
ggtitle("Boxplot of Time by Block")
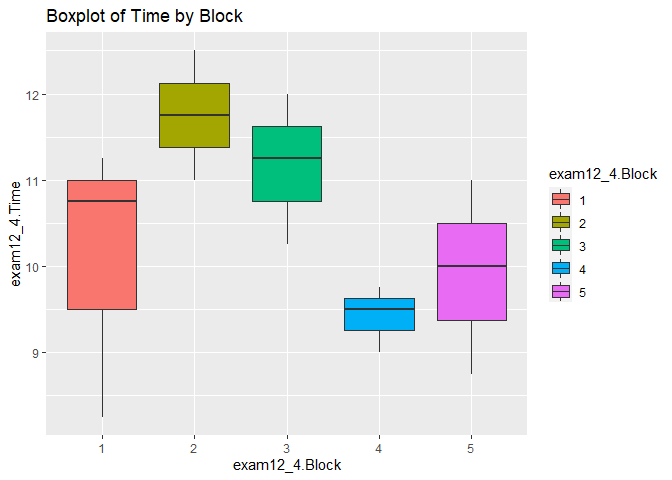
- 블록의 따른 마취 지속효과를 박스플롯으로 확인하여 보았다.
ggplot(ex12_4, aes(x = exam12_4.Treatment, y = exam12_4.Time, fill=exam12_4.Treatment)) +
geom_boxplot()+
ggtitle("Boxplot of Time by Treatment")
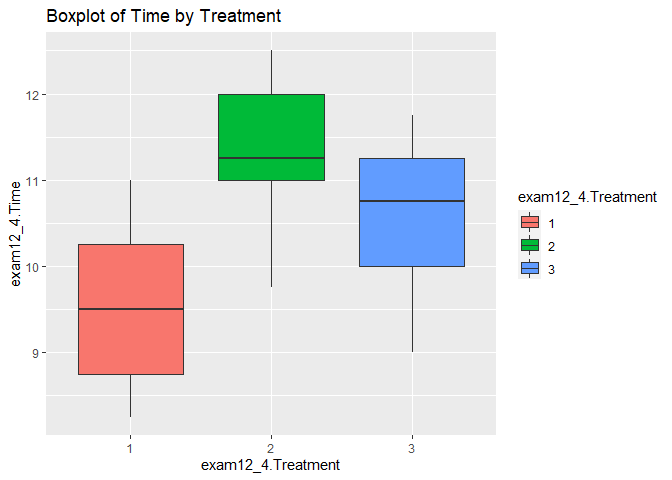
- 마취제의 따른 마취 지속효과를 박스플롯으로 확인하여 보았다.
Shapiro-Wilk Test로 정규성을 평가하였다.
shapiro.test(ex12_4$exam12_4.Time)
##
## Shapiro-Wilk normality test
##
## data: ex12_4$exam12_4.Time
## W = 0.97427, p-value = 0.9155
- p-value가 0.05보다 매우 크므로 모집단이 정규성을 따르지 않는다는 대립가설을 채택할 충분한 근거가 없다.
- 따라서 정규성을 만족한다는 사실을 바탕으로 등분산성 검정을 진행한다.
Bartlett’s Test를 통해 등분산성 검정을 진행하였다.
bartlett.test(ex12_4$exam12_4.Time~ex12_4$exam12_4.Treatment)
##
## Bartlett test of homogeneity of variances
##
## data: ex12_4$exam12_4.Time by ex12_4$exam12_4.Treatment
## Bartlett's K-squared = 0.01031, df = 2, p-value = 0.9949
- p-value가 모두 0.05보다 크므로 등분산성이 아니라는 아니라는 대립가설을 채택할 근거가 없다.
- 본 검정의 가설은 다음과 같다.
Randomized Complete Block ANOVA
aov(exam12_4.Time~exam12_4.Treatment+exam12_4.Block, data = ex12_4) %>%
tidy() %>%
rename(" "="term","Sum Sq"="sumsq","Mean Sq"="meansq","F value"="statistic","Pr(>F)"="p.value") %>%
kable(caption = "Randomized Complete Block ANOVA",booktabs = TRUE, valign = 't') %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| df | Sum Sq | Mean Sq | F value | Pr(\>F) | |
|---|---|---|---|---|---|
| exam12_4.Treatment | 2 | 7.708333 | 3.8541667 | 10.42254 | 0.0059166 |
| exam12_4.Block | 4 | 11.066667 | 2.7666667 | 7.48169 | 0.0082277 |
| Residuals | 8 | 2.958333 | 0.3697917 | NA | NA |
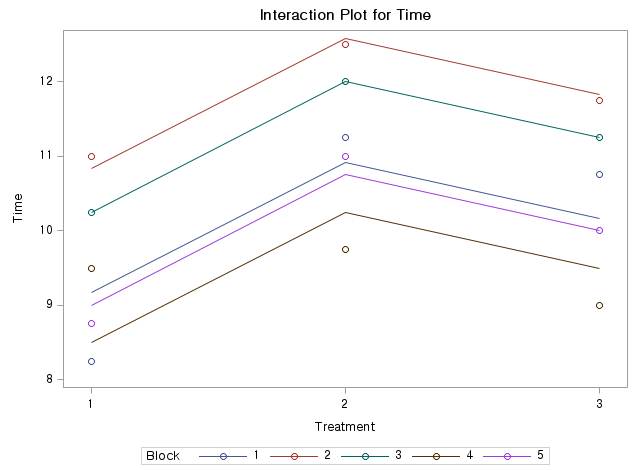
- 블록이 있는 분산분석을 수행한 결과이다.
- 분산분석 결과를 보면 Treatement와 블록의 p-value는 모두 0.05 보다 작으므로 귀무가설을 기각할 충분한 근거를 가지고 있다.
그러므로 마취약에 따라 마취의 효과 시간에 대한 모평균이 다르다라고 할 수 있다.
EXAMPLE 12.5
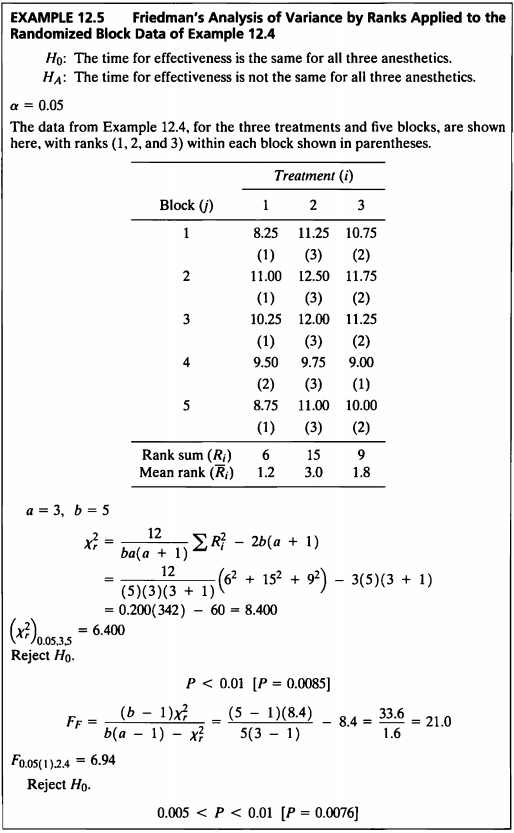
- Example 12.4의 데이터를 사용하여 비모수적 방법인 Friedman’s test를 수행하도록 한다.
- 이 검정 방법은 각 블록 내에 있는 Treatment 즉, 마취제의 효과가 있는 시간에 대한 평균을 가지고 Rank를 매긴다.
Friedman’s Test
library(agricolae)
fr <- friedman(ex12_4$exam12_4.Block, trt=ex12_4$exam12_4.Treatment, ex12_4$exam12_4.Time, alpha=0.05)
kable(fr[[1]], caption = paste0("Friedman's Test"))%>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"));kable(fr[[2]], caption = paste0("Parameters"))%>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"));kable(fr[[3]], caption = paste0("Means"))%>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"));kable(fr[[5]], caption = paste0("Post Hoc"))%>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| Chisq | Df | p.chisq | F | DFerror | p.F | t.value | LSD | |
|---|---|---|---|---|---|---|---|---|
| 8.4 | 2 | 0.0149956 | 21 | 8 | 0.0006554 | 2.306004 | 3.261182 |
| test | name.t | ntr | alpha | |
|---|---|---|---|---|
| Friedman | ex12_4$exam12_4.Treatment | 3 | 0.05 |
| ex12_4.exam12_4.Time | rankSum | std | r | Min | Max | Q25 | Q50 | Q75 |
|---|---|---|---|---|---|---|---|---|
| 9.55 | 6 | 1.109617 | 5 | 8.25 | 11.00 | 8.75 | 9.50 | 10.25 |
| 11.30 | 15 | 1.051784 | 5 | 9.75 | 12.50 | 11.00 | 11.25 | 12.00 |
| 10.55 | 9 | 1.081087 | 5 | 9.00 | 11.75 | 10.00 | 10.75 | 11.25 |
| Sum of ranks | groups | |
|---|---|---|
| 2 | 15 | a |
| 3 | 9 | b |
| 1 | 6 | b |
- 본 검정의 가설은 다음과 같다.
pf(as.numeric(fr$statistics[4]),2,4,lower.tail=F)
## [1] 0.007561437
- 프리드만 검정 결과 검정통계량 F값은 21로 나왔으며 p-value=0.0076으로 유의수준 0.05보다 작아 귀무가설을 기각할 충분한 근거를 가진다.
따라서 유의수준 5%하에 마취제에 따른 마취시간이 다르다고 할 수 있다.
EXAMPLE 12.6
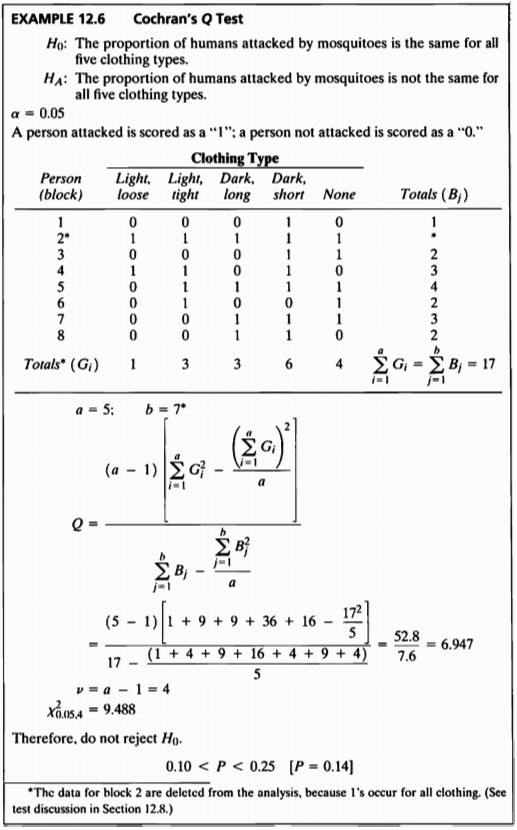
- 본 데이터는 5가지의 옷에 대해 모기에 공격을 당하는지 안당하는지를 기록한 데이터이다.
- Cochran’s Q test는 이항형으로 반복 관측된 데이터에 사용하는 방법으로 이항데이터에 대한 프리드만 검정이라고 할 수 있다.
ex12_6%>%
kbl(caption = "Dataset of Example 12.2") %>%
kable_styling(bootstrap_options = c("striped", "hover", "condensed", "responsive"))
| exam12_6.Person | exam12_6.Lightloose | exam12_6.Lighttight | exam12_6.Darklong | exam12_6.Darkshort | exam12_6.None |
|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 1 | 0 |
| 2 | 1 | 1 | 1 | 1 | 1 |
| 3 | 0 | 0 | 0 | 1 | 1 |
| 4 | 1 | 1 | 0 | 1 | 0 |
| 5 | 0 | 1 | 1 | 1 | 1 |
| 6 | 0 | 1 | 0 | 0 | 1 |
| 7 | 0 | 0 | 1 | 1 | 1 |
| 8 | 0 | 0 | 1 | 1 | 0 |
- 본 검정의 가설은 다음과 같다.
Cochran’s Test
library(nonpar)
ex12_6_del <- ex12_6[ex12_6$exam12_6.Person !=2,]
x = ex12_6_del[,-1]
cochrans.q(x)
##
## Cochran's Q Test
##
## H0: There is no difference in the effectiveness of treatments.
## HA: There is a difference in the effectiveness of treatments.
##
## Q = 6.94736842105263
##
## Degrees of Freedom = 4
##
## Significance Level = 0.05
## The p-value is 0.138695841622589
##
##
- 모든 종류의 옷을 입었을 때 모기에 물린 사람의 데이터는 삭제한다.
- Cochran’s Q Test를 수행한 결과, 검정통계량 값은 6.947, p-value는 0.14로 계산되었다.
- p-value가 0.05보다 크므로, 유의수준 5%하에 모기의 공격을 받은 사람의 비율은 옷의 종류에 따라 차이가 있다고 결론 내릴 수 없다.
SAS 프로그램 결과
SAS 접기/펼치기 버튼
12장
LIBNAME ex 'C:\Biostat';
RUN;
/*12장 연습문제 불러오기*/
%macro chap09(name=,no=);
%do i=1 %to &no.;
PROC IMPORT DBMS=excel
DATAFILE="C:\Biostat\data_chap12"
OUT=ex.&name.&i. REPLACE;
RANGE="exam12_&i.$";
RUN;
%end;
%mend;
%chap09(name=ex12_,no=6);
EXAMPLE 12.1
PROC SORT DATA=ex.ex12_1;
by sex;
run;
PROC MEANS DATA=ex.ex12_1 N Nmiss mean Sum;
class hormone;
by sex;
var plasma_calcium;
run;
PROC MEANS DATA=ex.ex12_1 N Nmiss mean Sum;
var plasma_calcium;
run;
MEANS 프로시저
| 분석 변수: plasma_calcium plasma_calcium | |||||
|---|---|---|---|---|---|
| Hormone | 관측값 수 | N | 결측값 수 | 평균 | 합계 |
| No Hormone | 5 | 5 | 0 | 14.8800000 | 74.4000000 |
| Yes Hormone | 5 | 5 | 0 | 32.5200000 | 162.6000000 |
| 분석 변수: plasma_calcium plasma_calcium | |||||
|---|---|---|---|---|---|
| Hormone | 관측값 수 | N | 결측값 수 | 평균 | 합계 |
| No Hormone | 5 | 5 | 0 | 12.1200000 | 60.6000000 |
| Yes Hormone | 5 | 5 | 0 | 27.7800000 | 138.9000000 |
MEANS 프로시저
| 분석 변수: plasma_calcium plasma_calcium | |||
|---|---|---|---|
| N | 결측값 수 | 평균 | 합계 |
| 20 | 0 | 21.8250000 | 436.5000000 |
EXAMPLE 12.2
/*정규성 검정*/
ods graphics off;ods exclude all;ods noresults;
PROC UNIVARIATE DATA=ex.ex12_1 normal plot;
CLASS hormone;
BY sex;
VAR plasma_calcium;
ODS OUTPUT TestsForNormality = TestsForNormality;
RUN;
ods graphics on;ods exclude none;ods results;
PROC SORT DATA=TestsForNormality;
BY descending Test;
RUN;
title " ex12_2 : 정규성 가정";
PROC PRINT DATA=TestsForNormality label;
WHERE Test="Shapiro-Wilk";
RUN;
title;
/*등분산성 검정*/
ods graphics off;ods exclude all;ods noresults;
PROC GLM DATA=ex.ex12_1;
CLASS hormone;
MODEL plasma_calcium=hormone/p;
MEANS hormone / HOVTEST=BARTLETT;
ODS OUTPUT Bartlett = Bartlett;
RUN;
PROC GLM DATA=ex.ex12_1;
CLASS sex;
MODEL plasma_calcium=sex/p;
MEANS sex / HOVTEST=BARTLETT;
ODS OUTPUT Bartlett = Bartlett2;
RUN;
ods graphics on;ods exclude none;ods results;
title "ex12_2 : 등분산 가정 (Bartlett's Test for Homogeneity of weight Variance)";
PROC PRINT DATA=Bartlett label;
RUN;
PROC PRINT DATA=Bartlett2 label;
RUN;
title;
| OBS | Sex | VarName | Hormone | 적합도 검정 | 적합도 통계량에 대한 레이블 | 적합도 통계량 값 | p-값 레이블 | p-값의 부호 | p-값 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | plasma_calcium | No Hormone | Shapiro-Wilk | W | 0.979256 | Pr < W | 0.9306 | |
| 2 | Female | plasma_calcium | Yes Hormone | Shapiro-Wilk | W | 0.982753 | Pr < W | 0.9488 | |
| 3 | Male | plasma_calcium | No Hormone | Shapiro-Wilk | W | 0.958824 | Pr < W | 0.7998 | |
| 4 | Male | plasma_calcium | Yes Hormone | Shapiro-Wilk | W | 0.978417 | Pr < W | 0.9260 |
| OBS | Effect | Dependent | Source | DF | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|
| 1 | Hormone | plasma_calcium | Hormone | 1 | 0.1999 | 0.6548 |
| OBS | Effect | Dependent | Source | DF | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|
| 1 | Sex | plasma_calcium | Sex | 1 | 0.0915 | 0.7623 |
EXAMPLE 12.3
PROC GLM DATA=ex.ex12_1;
CLASS Hormone sex;
MODEL plasma_calcium=Hormone sex Hormone*sex;
LSMEANS Hormone sex/diff CL adjust=tukey;
ODS OUTPUT LSMeanDiffCL=LSMeanDiffCL;
RUN;
The GLM Procedure
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Hormone | 2 | No Hormone Yes Hormone |
| Sex | 2 | Female Male |
| Number of Observations Read | 20 |
|---|---|
| Number of Observations Used | 20 |
The GLM Procedure
Dependent Variable: plasma_calcium plasma_calcium
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Model | 3 | 1461.325500 | 487.108500 | 25.86 | <.0001 |
| Error | 16 | 301.392000 | 18.837000 | ||
| Corrected Total | 19 | 1762.717500 |
| R-Square | Coeff Var | Root MSE | plasma_calcium Mean |
|---|---|---|---|
| 0.829019 | 19.88619 | 4.340161 | 21.82500 |
| Source | DF | Type I SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Hormone | 1 | 1386.112500 | 1386.112500 | 73.58 | <.0001 |
| Sex | 1 | 70.312500 | 70.312500 | 3.73 | 0.0713 |
| Hormone*Sex | 1 | 4.900500 | 4.900500 | 0.26 | 0.6170 |
| Source | DF | Type III SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Hormone | 1 | 1386.112500 | 1386.112500 | 73.58 | <.0001 |
| Sex | 1 | 70.312500 | 70.312500 | 3.73 | 0.0713 |
| Hormone*Sex | 1 | 4.900500 | 4.900500 | 0.26 | 0.6170 |
The GLM Procedure
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
| Hormone | plasma_calcium LSMEAN | H0:LSMean1=LSMean2 |
|---|---|---|
| Pr > |t| | ||
| No Hormone | 13.5000000 | <.0001 |
| Yes Hormone | 30.1500000 |
| Hormone | plasma_calcium LSMEAN | 95% Confidence Limits | |
|---|---|---|---|
| No Hormone | 13.500000 | 10.590473 | 16.409527 |
| Yes Hormone | 30.150000 | 27.240473 | 33.059527 |
| Least Squares Means for Effect Hormone | ||||
|---|---|---|---|---|
| i | j | Difference Between Means | Simultaneous 95% Confidence Limits for LSMean(i)-LSMean(j) | |
| 1 | 2 | -16.650000 | -20.764501 | -12.535499 |
The GLM Procedure
Least Squares Means
Adjustment for Multiple Comparisons: Tukey
| Sex | plasma_calcium LSMEAN | H0:LSMean1=LSMean2 |
|---|---|---|
| Pr > |t| | ||
| Female | 23.7000000 | 0.0713 |
| Male | 19.9500000 |
| Sex | plasma_calcium LSMEAN | 95% Confidence Limits | |
|---|---|---|---|
| Female | 23.700000 | 20.790473 | 26.609527 |
| Male | 19.950000 | 17.040473 | 22.859527 |
| Least Squares Means for Effect Sex | ||||
|---|---|---|---|---|
| i | j | Difference Between Means | Simultaneous 95% Confidence Limits for LSMean(i)-LSMean(j) | |
| 1 | 2 | 3.750000 | -0.364501 | 7.864501 |
PROC PRINT DATA=LSMeanDiffCL;
RUN;
| OBS | Effect | Dependent | i | j | LowerCL | Difference | UpperCL | Hormone | _Hormone | Sex | _Sex |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Hormone | plasma_calcium | 1 | 2 | -20.764501 | -16.650000 | -12.535499 | No Hormone | Yes Hormone | ||
| 2 | Sex | plasma_calcium | 1 | 2 | -0.364501 | 3.750000 | 7.864501 | Female | Male |
PROC IML;
USE ex.ex12_1;
READ all;
CLOSE ex.ex12_1;
female=loc(Sex = "Female");
male=loc(Sex = "Male");
total_F = sum(plasma_calcium[female,]);
total_M = sum(plasma_calcium[male,]);
number_F = nrow(plasma_calcium[female,]);
number_M= nrow(plasma_calcium[male,]);
Xbar_p = round((total_F + total_M) / (number_F + number_M), 0.001);
t_quantile = round(quantile('T', 1-0.05/2, 16), 0.001);
s2 = 18.8370;
margin = t_quantile * sqrt(s2/(number_F+number_M));
lower_bound = round(Xbar_p - margin, 0.001) ;
upper_bound = round(Xbar_p + margin, 0.001) ;
*print female male;
*print total_F total_M number_F number_M;
PRINT Xbar_p lower_bound upper_bound ;
RUN;
QUIT;
| Xbar_p | lower_bound | upper_bound |
|---|---|---|
| 21.825 | 19.768 | 23.882 |
EXAMPLE 12.4
/*정규성 검정*/
ods graphics off;ods exclude all;ods noresults;
PROC UNIVARIATE DATA=ex.ex12_4 normal plot;
VAR Time;
ODS OUTPUT TestsForNormality = TestsForNormality;
RUN;
ods graphics on;ods exclude none;ods results;
PROC SORT DATA=TestsForNormality;
BY descending Test;
RUN;
title " ex12_2 : 정규성 가정";
PROC PRINT DATA=TestsForNormality label;
WHERE Test="Shapiro-Wilk";
RUN;
title;
/*등분산성 검정*/
ods graphics off;ods exclude all;ods noresults;
PROC GLM DATA=ex.ex12_4;
CLASS Treatment;
MODEL Time=Treatment/p;
MEANS Treatment / HOVTEST=BARTLETT;
ODS OUTPUT Bartlett = Bartlett;
RUN;
ods graphics on;ods exclude none;ods results;
title "ex12_2 : 등분산 가정 (Bartlett's Test for Homogeneity of weight Variance)";
PROC PRINT DATA=Bartlett label;
RUN;
title;
| OBS | VarName | 적합도 검정 | 적합도 통계량에 대한 레이블 | 적합도 통계량 값 | p-값 레이블 | p-값의 부호 | p-값 |
|---|---|---|---|---|---|---|---|
| 1 | Time | Shapiro-Wilk | W | 0.974271 | Pr < W | 0.9155 |
| OBS | Effect | Dependent | Source | DF | Chi-Square | Pr > ChiSq |
|---|---|---|---|---|---|---|
| 1 | Treatment | Time | Treatment | 2 | 0.0103 | 0.9949 |
ods graphics off;ods exclude all;ods noresults;
PROC GLM DATA=ex.ex12_4;
CLASS Treatment Block;
MODEL Time=Treatment Block;
RANDOM block;
RUN;
ods graphics on;ods exclude none;ods results;
title "Example12.4 ";
ods graphics off;ods exclude all;ods noresults;
PROC GLM DATA=ex.ex12_4 plots=(DIAGNOSTICS RESIDUALS);
CLASS Treatment Block;
MODEL Time = Treatment Block;
RANDOM Block / test;
ODS OUTPUT OverallANOVA = OverallANOVA_4
RandomModelANOVA = RandomModelANOVA_4;
RUN;
ods graphics on;ods exclude none;ods results;
PROC PRINT DATA=RandomModelANOVA_4 label;
VAR Source DF SS MS FValue ProbF;
RUN;
| OBS | Source | DF | Type III SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|---|
| 1 | Treatment | 2 | 7.708333 | 3.854167 | 10.42 | 0.0059 |
| 2 | Block | 4 | 11.066667 | 2.766667 | 7.48 | 0.0082 |
| 3 | Error: MS(Error) | 8 | 2.958333 | 0.369792 | _ | _ |
EXAMPLE 12.5
title "Example12.5 ";
PROC SORT DATA=ex.ex12_4;
BY block;
RUN;
PROC RANK DATA=ex.ex12_4 out=ex12_4;
BY Block;
VAR time;
RANKS Rtime;
RUN;
PROC PRINT;
title2 'Original and Ranked Values of Yield';
RUN;
PROC ANOVA DATA=ex12_4;
CLASS Block Treatment;
MODEL Rtime = Block Treatment;
title2 'Friedman''s Two-way Nonparametric ANOVA';
RUN;
| OBS | Treatment | Block | Time | Rtime |
|---|---|---|---|---|
| 1 | 1 | 1 | 8.25 | 1 |
| 2 | 2 | 1 | 11.25 | 3 |
| 3 | 3 | 1 | 10.75 | 2 |
| 4 | 1 | 2 | 11.00 | 1 |
| 5 | 2 | 2 | 12.50 | 3 |
| 6 | 3 | 2 | 11.75 | 2 |
| 7 | 1 | 3 | 10.25 | 1 |
| 8 | 2 | 3 | 12.00 | 3 |
| 9 | 3 | 3 | 11.25 | 2 |
| 10 | 1 | 4 | 9.50 | 2 |
| 11 | 2 | 4 | 9.75 | 3 |
| 12 | 3 | 4 | 9.00 | 1 |
| 13 | 1 | 5 | 8.75 | 1 |
| 14 | 2 | 5 | 11.00 | 3 |
| 15 | 3 | 5 | 10.00 | 2 |
The ANOVA Procedure
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| Block | 5 | 1 2 3 4 5 |
| Treatment | 3 | 1 2 3 |
| Number of Observations Read | 15 |
|---|---|
| Number of Observations Used | 15 |
The ANOVA Procedure
Dependent Variable: Rtime 변수 Time의 순위
| Source | DF | Sum of Squares | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Model | 6 | 8.40000000 | 1.40000000 | 7.00 | 0.0075 |
| Error | 8 | 1.60000000 | 0.20000000 | ||
| Corrected Total | 14 | 10.00000000 |
| R-Square | Coeff Var | Root MSE | Rtime Mean |
|---|---|---|---|
| 0.840000 | 22.36068 | 0.447214 | 2.000000 |
| Source | DF | Anova SS | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|
| Block | 4 | 0.00000000 | 0.00000000 | 0.00 | 1.0000 |
| Treatment | 2 | 8.40000000 | 4.20000000 | 21.00 | 0.0007 |
EXAMPLE 12.6
title "Example12.6 ";
DATA ex.ex12_6;
SET ex.ex12_6;
IF person=2 THEN delete;
RUN;
ods graphics off;ods exclude all;ods noresults;
PROC FREQ DATA=ex.ex12_6;
TABLES Lightloose Lighttight Darklong Darkshort None / nocum;
TABLES Lightloose*Lighttight*Darklong*Darkshort*None / agree noprint;
ODS OUTPUT CochransQ = CochransQ;
RUN;
ods graphics on;ods exclude none;ods results;
PROC PRINT DATA=CochransQ;
RUN;
| OBS | Table | ChiSquare | DF | Prob |
|---|---|---|---|---|
| 1 | Darkshort * None에 대한 요약 | 6.9474 | 4 | 0.1387 |
교재: Biostatistical Analysis (5th Edition) by Jerrold H. Zar
**이 글은 22학년도 1학기 의학통계방법론 과제 자료들을 정리한 글 입니다.**
